Lead Image: Researchers from UNC School of Medicine are using evolutionary genomics to understand human diseases. They focus on unchanged, highly constrained genes through the mammalian evolution. This new perspective allows tracing of psychiatric and neurological disorders to alterations in these genes. This could provide insights into various mental health conditions and possibly allow researchers to lessen the effects of genetic diseases by manipulating certain DNA sequences.
A team of scientists has created a new manual researchers can use to learn more about the origins of human diseases with high genetic risk.
Hundreds of scientific studies have been conducted over the years to find the genes underlying common human traits, from eye color to intelligence and physical and mental illnesses.
Patrick Sullivan, MD, FRANZCP, the Yeargan Distinguished Professor of Psychiatry and Genetics at the University of North Carolina (UNC) School of Medicine, and the Psychiatric Genomic Consortium have produced a new packet for the journal Science, to give researchers another way to understand human disease, using the power of evolutionary genomics.
“This is a tool that can give us a lot of important hints about human disease,” said Sullivan, who is also a professor at the Karolinska Institute in Stockholm, Sweden. “If we can take a deep dive into your genome, we can get some idea about your ancestors, both human and nonhuman, and observe the impacts of many millions of years of evolution in you.”
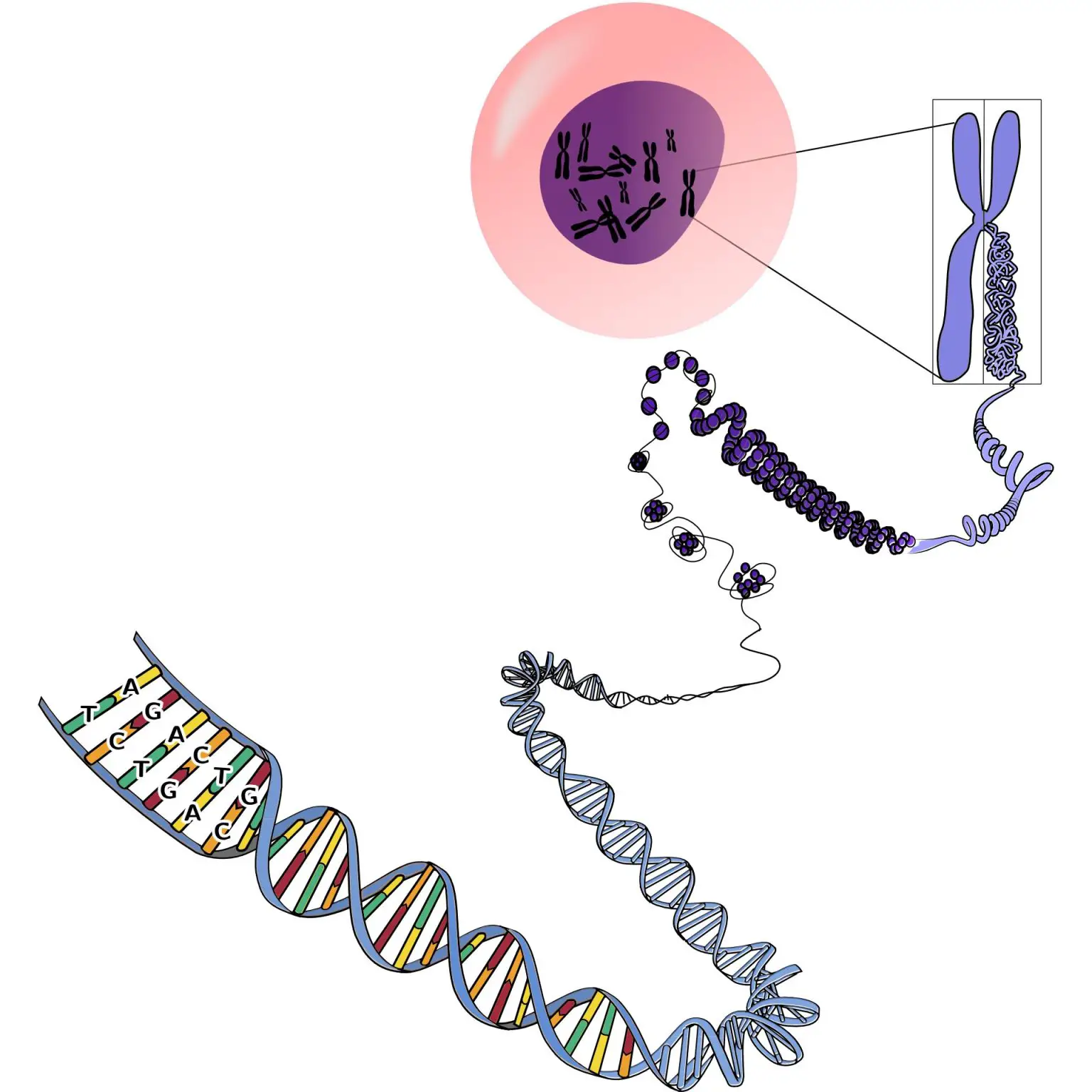
What Makes Us Mammals
Every single living organism on the planet has DNA. The self-replicating material acts as a blueprint for producing certain molecules in organisms, such as proteins. It’s no surprise that humans and our closest relatives, chimpanzees, share 98.8% of genetic material.
While some of our genes have evolved over time, others have remained the same throughout the entire mammalian evolutionary process. In scientific terms, these are called “highly constrained” genes. Some human genes have a surprising amount of genetic similarity in mice, cows, dogs, cats, bats, and dolphins in many regions of the genome.
These are the genes that unite us as mammals. Since these genes have undergone a “trial by fire” throughout evolutionary history, these unaltered genetic regions must play a fundamental role in the health and genetic makeup of the organism, according to Sullivan.

“Some highly constrained genes can make proteins that are nearly identical in us and in a mouse,” said Sullivan. “That’s crazy because we have probably 60 million years of evolution between us and the mouse. And yet, this protein hasn’t changed so we infer that this protein is doing something really important.”
It might be simpler to see the work of our shared genes when we zoom out to take a more holistic view.
Humans and other mammals share anatomical structures, such as the four-chambered heart, lungs, hair (or fur), skeleton, and milk-producing mammary glands. We also share similar fundamental processes on a smaller scale, including embryology, how cells grow and divide, and the development and operation of the synapses that transmit neurological chemicals throughout our bodies and brains.
All of which are formed through our shared genetic regions. So, if one of these genes that make up the basics of a mammal is altered or deleted, it could have negative effects on the organism.
A New Way to Look at Human Mental and Physical Health
If a patient has a neurological brain disorder or certain psychiatric disorders, researchers are able to trace it back and see that this person has received a “big hit” to one of the highly constrained genes that are critical to the nervous system, brain structure, or synapses.
Many researchers have relied on the genome-wide association study (GWAS) to find where the genetic risk for a disease is located in the genome. Using genomic techniques and large-size samples, researchers can analyze the entire genome of many populations to find genetic variations, such as single nucleotide polymorphisms (SNPs), associated with a disease or a trait.
Even though it is important to know where these variations are located in the genome, it’s also useful to know how or why these genetic variations happened in the first place. Sullivan hopes that other researchers will make use of the new and extensive document to reach their own conclusions regarding the genetics underlying a variety of human diseases.
“As it turns out, a lot of brain traits are actually highly conserved,” said Sullivan, who serves as director of the UNC Suicide Prevention Institute in the Department of Psychiatry. “This research project has really given me a much, much deeper understanding of the genome and how the genome is set up. I now use this all the time in trying to understand schizophrenia, suicide, depression, and eating disorders.”
What This Means for Future Research
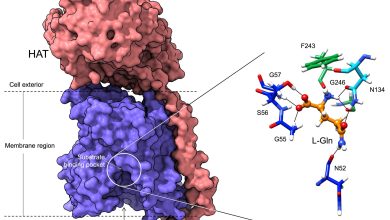
As one can imagine, the successful development of a human requires heavy lifting from proteins and DNA sequences. There are two short regions within our DNA, called regulatory enhancers and regulatory promoters, which play especially important roles in regulating our DNA.
The creation of a human gene is similar to a factory that produces donuts. Regulatory enhancers are responsible for controlling the amount of dough squeezed out of the machine and onto the baking tray. Promoters, on the other hand, are in control of when the dough is being squirted onto the tray. At the end of the day, you have a full formed gene.
Researchers like Sullivan may be able to go into the DNA sequences and increase or decrease these regulatory enhancers and promoters to affect the amount of proteins produced by genes, with the goal of lessening the effects of a genetically based disease.
“It might be possible to hit the upstream part that controls it, in a very soft way, to see if that actually helps,” says Sullivan.
Reference: “Leveraging base-pair mammalian constraint to understand genetic variation and human disease” by Patrick F. Sullivan, Jennifer R. S. Meadows, Steven Gazal, BaDoi N. Phan, Xue Li, Diane P. Genereux, Michael X. Dong, Matteo Bianchi, Gregory Andrews, Sharadha Sakthikumar, Jessika Nordin, Ananya Roy, Matthew J. Christmas, Voichita D. Marinescu, Chao Wang, Ola Wallerman, James Xue, Shuyang Yao, Quan Sun, Jin Szatkiewicz, Jia Wen, Laura M. Huckins, Alyssa Lawler, Kathleen C. Keough, Zhili Zheng, Jian Zeng, Naomi R. Wray, Yun Li, Jessica Johnson, Jiawen Chen, Zoonomia Consortium§. , Benedict Paten, Steven K. Reilly, Graham M. Hughes, Zhiping Weng, Katherine S. Pollard, Andreas R. Pfenning, Karin Forsberg-Nilsson, Elinor K. Karlsson, Kerstin Lindblad-Toh, Gregory Andrews, Joel C. Armstrong, Matteo Bianchi, Bruce W. Birren, Kevin R. Bredemeyer, Ana M. Breit, Matthew J. Christmas, Hiram Clawson, Joana Damas, Federica Di Palma, Mark Diekhans, Michael X. Dong, Eduardo Eizirik, Kaili Fan, Cornelia Fanter, Nicole M. Foley, Karin Forsberg-Nilsson, Carlos J. Garcia, John Gatesy, Steven Gazal, Diane P. Genereux, Linda Goodman, Jenna Grimshaw, Michaela K. Halsey, Andrew J. Harris, Glenn Hickey, Michael Hiller, Allyson G. Hindle, Robert M. Hubley, Graham M. Hughes, Jeremy Johnson, David Juan, Irene M. Kaplow, Elinor K. Karlsson, Kathleen C. Keough, Bogdan Kirilenko, Klaus-Peter Koepfli, Jennifer M. Korstian, Amanda Kowalczyk, Sergey V. Kozyrev, Alyssa J. Lawler, Colleen Lawless, Thomas Lehmann, Danielle L. Levesque, Harris A. Lewin, Xue Li, Abigail Lind, Kerstin Lindblad-Toh, Ava Mackay-Smith, Voichita D. Marinescu, Tomas Marques-Bonet, Victor C. Mason, Jennifer R. S. Meadows, Wynn K. Meyer, Jill E. Moore, Lucas R. Moreira, Diana D. Moreno-Santillan, Kathleen M. Morrill, Gerard Muntané, William J. Murphy, Arcadi Navarro, Martin Nweeia, Sylvia Ortmann, Austin Osmanski, Benedict Paten, Nicole S. Paulat, Andreas R. Pfenning, BaDoi N. Phan, Katherine S. Pollard, Henry E. Pratt, David A. Ray, Steven K. Reilly, Jeb R. Rosen, Irina Ruf, Louise Ryan, Oliver A. Ryder, Pardis C. Sabeti, Daniel E. Schäffer, Aitor Serres, Beth Shapiro, Arian F. A. Smit, Mark Springer, Chaitanya Srinivasan, Cynthia Steiner, Jessica M. Storer, Kevin A. M. Sullivan, Patrick F. Sullivan, Elisabeth Sundström, Megan A. Supple, Ross Swofford, Joy-El Talbot, Emma Teeling, Jason Turner-Maier, Alejandro Valenzuela, Franziska Wagner, Ola Wallerman, Chao Wang, Juehan Wang, Zhiping Weng, Aryn P. Wilder, Morgan E. Wirthlin, James R. Xue and Xiaomeng Zhang, 28 April 2023, Science.
DOI: 10.1126/science.abn2937