File Manager :
Hey guys,
This week I am back with a review for file manager app. It is named File Expert – File Manager.
It is available here: https://play.google.com/store/apps/details?id=xcxin.filexpert&hl=en
As we all know, there is a lot of different File Manager apps on the Google Play store and it is pretty hard to differentiate when making this kind of app. Did this app solve that challenge? Is it a good app or not? We will analyze the features and UI and give the first even Jon’s Grade. Let’s do this!
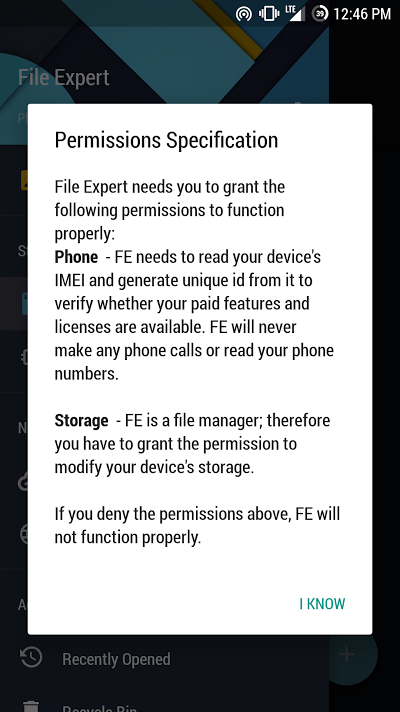 The first thing you will notice after installing this app, is that it explains the permissions it will need before it asks you for them. That is one thing that I hve never seen before and I truly appreciated.
The first thing you will notice after installing this app, is that it explains the permissions it will need before it asks you for them. That is one thing that I hve never seen before and I truly appreciated.
Next, if you go through the side menu (which you can access by swiping from the left side of the phone, you will see a pretty standard menu when it comes to file managers. The app comes with the options of which storage to go to. Internal storage was named SD Card which is reminiscent of the old Android OS. I personally did not like it. When you put in a SD card, you have 2 SD cards in there. Contrary to the other File Manager I use that just labels it Internal Storage.
Next you have network options which later also became pretty standard. Advanced features provided Recently Opened and Tags which were different to what I’m used to and very nice to have. The Recycle Bin is standard too but it’s such an important feature to me by now. Having the ability to delete something and recover it for the Recycle Bin is amazing. It’s hardly new since it’s the same as any computers, but the early file managers on Android didn’t have that and you had to delete stuff permanently.
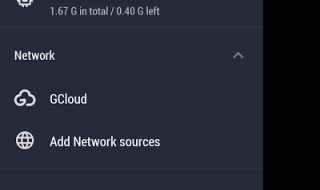
Next, you can find your files by Gallery, Music, Video, Apps, Compressed and Documents. Convenient. They also have tools. While I’ve seen the Downloader and FTP Server before, the Safebox is somewhat new to me. So I went in and decided to try it out. They let me set a pattern password and asked for an email in case I forgot the pattern. Then, when I went to copy/paste a file to test it, I was prompted to pay for a premium account to use the feature. I am sure it is worth the money, but I am only reviewing the free app here so we will have to pass. I do have to say, I find it very expensive for the few features a premium account gives me. I have these features for free with other apps.
All in all, the app performs well and the huge plus is the material design. WOW. It is a very beautiful app to work with when it comes to UI and navigation. It is simple and performs normal file manager tasks very easily.

This is my first app review on TekhDecoded and I have decided to give apps I review a Jon’s Grade. It will be on a scale of 1 to 10 and will take into account several things like features, UI, usability and cost of premium features.
So this app receives a Jon’s Grade of 7/10. The overall package is amazing looking and easy to use. It only loses points when it comes to what is considered premium features and the price for those.
If you are in the lookout for a File Manager, I would recommend this with no problem.
Their paid features have 30 days free trial period, you can buy them and cancel within 30 days. Do try them if you want a clean experience of yourself to love this app! 😀
Let me know in the comments below if you agree or not!http://www.tbcredit.ru/zaymyi.html