
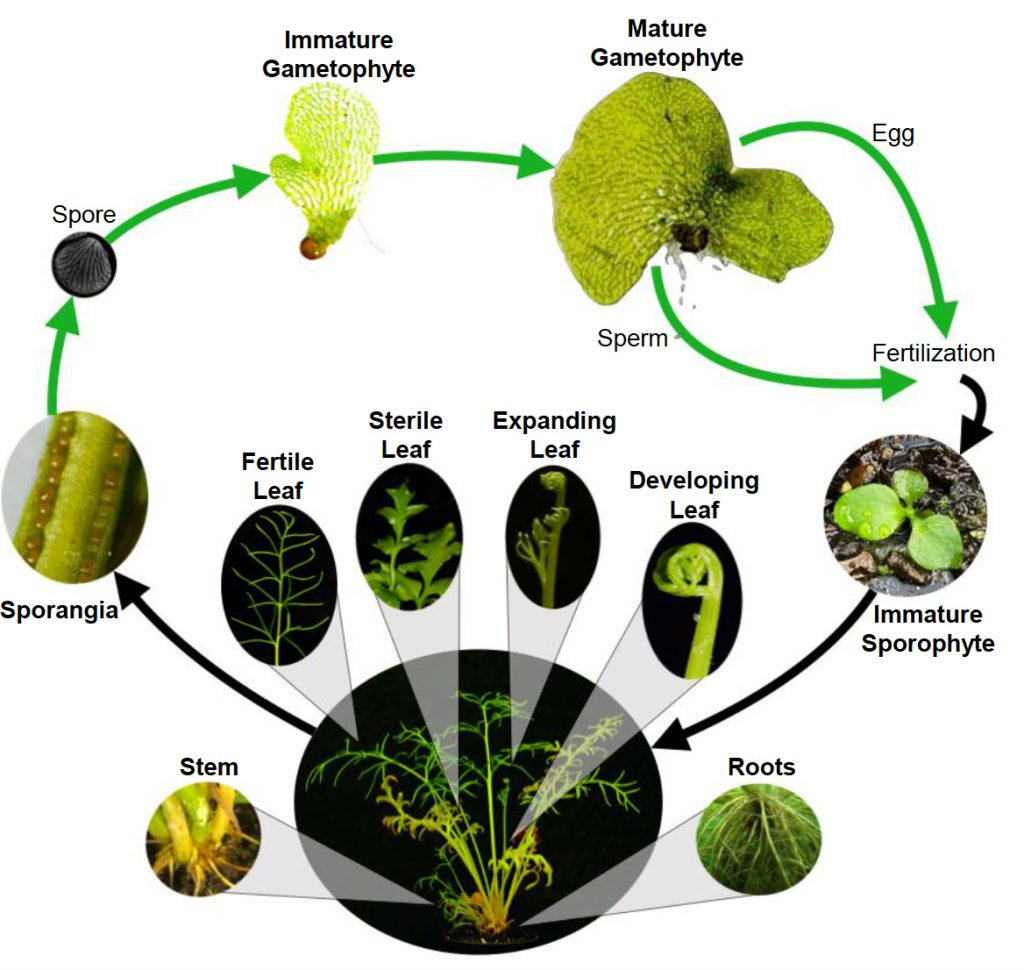
Lead Image: Ferns are vascular plants that reproduce through spores and do not have seeds or flowers.
A new study reveals ferns’ history of DNA hoarding and kleptomania.
Ferns are infamous for having an enormous number of chromosomes and massive amounts of DNA. A fern no larger than a dinner plate currently holds the record for highest chromosome count, with 720 pairs packed into each of its nuclei. Scientists have been baffled by ferns’ tendency to hoard DNA, and the intractable size of their genomes has made it challenging to sequence, assemble, and interpret them.
Now, two articles recently published in the journal Nature Plants are rewriting history with the first full-length genomes for homosporous ferns, a huge group that encompasses 99% of all modern fern diversity.
“Every genome tells a different story,” said co-author Doug Soltis, a distinguished professor with the Florida Museum of Natural History. “Ferns are the closest living relatives of all seed plants, and they produce chemical deterrents to herbivores that may be useful for agricultural research. Yet until now, they’ve remained the last major lineage of green life without a genome sequence.”

Recently, two different research teams independently published the genomes of the flying spider monkey tree fern (Alsophila spinulosa) and Ceratopteris (Ceratopteris richardii).
The Ceratopteris genome analysis provides hints for answering the long-standing puzzle of why ferns store more DNA than other plants on average. Comparisons to other species’ genomes revealed that ferns stole the genes for some of their anti-herbivory toxins from bacteria.
The Ceratopteris genome bucks a decades-old theory, leaving more questions than answers
Since the 1960s, the most favored explanation for why ferns contain so much DNA invoked rampant whole-genome duplications, in which an extra set of chromosomes is accidentally passed on to an organism’s offspring. This can sometimes be beneficial, as all the extra genes can then be used as raw material for the evolution of new traits. In fact, whole-genome duplication has been implicated in the origin of nearly all crop plants.
Whole-genome duplication is common in plants and even some animals, but most organisms tend to jettison the extra genetic baggage over time, slimming back down to smaller genomes that are metabolically easier to maintain.
“This has been a major point of discussion for the last half-century and has led to all kinds of conflicting results,” said lead author Blaine Marchant, a postdoctoral scholar at Stanford University and former Florida Museum graduate student. “Trying to figure out the evolutionary process underlying this paradox is incredibly important.”
With the first fully assembled homosporous fern genomes, scientists were finally prepared to address this question, but getting there wasn’t easy. Sequencing the large, complex genome of Ceratopteris took over eight years of work and the combined effort of dozens of researchers from 28 institutions around the world, including the U.S. Department of Energy Joint Genome Institute. The final result was 7.46 gigabases of DNA, more than double the size of the human genome.
If Ceratopteris had bulked up on DNA through repeated genome duplication events, researchers expected large portions of its 39 chromosome pairs would be identical. What they found instead was a mixed bag of repetitive sequences and millions of short snippets called jumping genes, which accounted for 85% of the fern’s DNA. Rather than multiple genome copies, Ceratopteris mostly contains genetic debris accumulated over millions of years.
“The functional genes are separated by large amounts of repetitive DNA. And although we’re not yet sure how the Ceratopteris and other fern genomes got so big, it’s clear that the prevailing view of repeated episodes of genome duplication is not supported,” said co-author Pam Soltis, a Florida Museum curator and distinguished professor.
The authors note that it’s too early to make any firm conclusions, especially since this is the first analysis of its scope conducted in this group. Cross comparisons with additional fern genomes down the road will help paint a clearer picture of how these plants evolved.
Still, the results point to a clear difference in the way homosporous ferns manage their genetic content compared to almost all other plants, Marchant said.
“What we seem to be finding is that things like flowering plants, which on average have much smaller genomes than ferns, are just better at getting rid of junk DNA. They’re better at dropping spare chromosomes and even downsizing after small duplications.”
Ferns repeatedly stole toxins from bacteria
A closer look at the billions of DNA base pairs within Ceratopteris revealed multiple defense genes that code for a particularly sinister type of pore-forming toxin. These toxins bind to cells, where they become activated and form small, hollow rings that punch their way into the cellular membrane. Water floods into the cells through the resulting holes, causing them to rupture.
Pore-forming toxins have been intensively studied by scientists for their potential use in nanopore technology, Marchant explained. Most often, however, they’re found in bacteria.
“This is the first concrete evidence of these bacterial toxin-related genes within fern DNA,” Marchant said, noting that the similarity isn’t a coincidence.
Rather than evolving this toxin on its own, Ceratopteris appears to have obtained it directly from bacteria through a process called horizontal gene transfer. And given that there were multiple copies of the gene spread out among three separate chromosomes, it’s likely this happened more than once.
“What’s fascinating is that the many copies of these genes show up in different parts of the plant,” he said. “Some are highly expressed in the stem and roots, while other copies are expressed solely in the leaves, and others are generally expressed across all tissues. We cannot be sure of the exact function of these genes at this point, but their similarity to the toxin-forming genes in bacteria certainly suggests these genes are defense-related.”
This wouldn’t be the first time ferns have incorporated foreign DNA into their genomes. A 2014 study indicates ferns may have evolved their characteristic ability to grow in shady environments by borrowing genes from distantly related plants.
However, exactly how organisms separated by millions of years of evolution are able to swap fully functional genes remains unclear.
“The mechanisms behind horizontal gene transfer remain one of the least investigated areas of land plant evolution,” Doug Soltis explained. “Over evolutionary timescales, it’s a bit like winning the lottery. Any time a plant is wounded, its interior is susceptible to invasion from microbes, but for their DNA to be incorporated into the genome seems amazing.”
The authors say this is merely the first step in a long series of studies with practical applications ranging from the development of novel biopesticides to innovative new conservation strategies.
References:
“Dynamic genome evolution in a model fern” by D. Blaine Marchant, Guang Chen, Shengguan Cai, Fei Chen, Peter Schafran, Jerry Jenkins, Shengqiang Shu, Chris Plott, Jenell Webber, John T. Lovell, Guifen He, Laura Sandor, Melissa Williams, Shanmugam Rajasekar, Adam Healey, Kerrie Barry, Yinwen Zhang, Emily Sessa, Rijan R. Dhakal, Paul G. Wolf, Alex Harkess, Fay-Wei Li, Clemens Rössner, Annette Becker, Lydia Gramzow, Dawei Xue, Yuhuan Wu, Tao Tong, Yuanyuan Wang, Fei Dai, Shuijin Hua, Hua Wang, Shengchun Xu, Fei Xu, Honglang Duan, Günter Theißen, Michael R. McKain, Zheng Li, Michael T. W. McKibben, Michael S. Barker, Robert J. Schmitz, Dennis W. Stevenson, Cecilia Zumajo-Cardona, Barbara A. Ambrose, James H. Leebens-Mack, Jane Grimwood, Jeremy Schmutz, Pamela S. Soltis, Douglas E. Soltis and Zhong-Hua Chen, 1 September 2022, Nature Plants.
DOI: 10.1038/s41477-022-01226-7
“The flying spider-monkey tree fern genome provides insights into fern evolution and arborescence” by Xiong Huang, Wenling Wang, Ting Gong, David Wickell, Li-Yaung Kuo, Xingtan Zhang, Jialong Wen, Hoon Kim, Fachuang Lu, Hansheng Zhao, Song Chen, Hui Li, Wenqi Wu, Changjiang Yu, Su Chen, Wei Fan, Shuai Chen, Xiuqi Bao, Li Li, Dan Zhang, Longyu Jiang, Xiaojing Yan, Zhenyang Liao, Gongke Zhou, Yalong Guo, John Ralph, Ronald R. Sederoff, Hairong Wei, Ping Zhu, Fay-Wei Li, Ray Ming and Quanzi Li, 9 May 2022, Nature Plants.
DOI: 10.1038/s41477-022-01146-6
“Horizontal transfer of an adaptive chimeric photoreceptor from bryophytes to ferns” by Fay-Wei Li, Juan Carlos Villarreal, Steven Kelly, Carl J. Rothfels, Michael Melkonian, Eftychios Frangedakis, Markus Ruhsam, Erin M. Sigel, Joshua P. Der, Jarmila Pittermann, Dylan O. Burge, Lisa Pokorny, Anders Larsson, Tao Chen, Stina Weststrand, Philip Thomas, Eric Carpenter, Yong Zhang, Zhijian Tian, Li Chen, Zhixiang Yan, Ying Zhu, Xiao Sun, Jun Wang, Dennis W. Stevenson, Barbara J. Crandall-Stotler, A. Jonathan Shaw, Michael K. Deyholos, Douglas E. Soltis, Sean W. Graham, Michael D. Windham, Jane A. Langdale, Gane Ka-Shu Wong, Sarah Mathews and Kathleen M. Pryer, 14 April 2014, Proceedings of the National Academy of Sciences.
DOI: 10.1073/pnas.1319929111
Several of the authors are involved in the current effort to sequence the genomes of all known eukaryotic organisms within a 10-year time frame. Called the Earth Biogenome Project, the endeavor will generate untold genomic resources that researchers will have their hands full analyzing for the foreseeable future.
The study was funded by the National Science Foundation, the National Natural Science Foundation of China, the Australian Research Council, Horticulture Innovation Australia, the Ambrose Monell Foundation, the Key R&D Program of Zhejiang Province, the Zhejiang Provincial Natural Science Foundation of China, and the China Agriculture Research System.





