MIT researchers have found a way to store quantum information in the vibrational motion of atom pairs, similar to the swinging motion of two pendula, connected by a spring. The quantum register contains hundreds of pairs of vibrating qubits that researchers can coherently control for over ten seconds. Credit: Sampson Wilcox/RLE
The new qubits stay in “superposition” for up to 10 seconds, and could make a promising foundation for quantum computers.
MIT physicists have discovered a new quantum bit, or “qubit,” in the form of vibrating pairs of atoms known as fermions. They found that when pairs of fermions are chilled and trapped in an optical lattice, the particles can exist simultaneously in two states — a weird quantum phenomenon known as superposition. In this case, the atoms held a superposition of two vibrational states, in which the pair wobbled against each other while also swinging in sync, at the same time.
The team was able to maintain this state of superposition among hundreds of vibrating pairs of fermions. In so doing, they achieved a new “quantum register,” or system of qubits, that appears to be robust over relatively long periods of time. The discovery, published on January 26, 2022, in the journal Nature, demonstrates that such wobbly qubits could be a promising foundation for future quantum computers.
A qubit represents a basic unit of quantum computing. Where a classical bit in today’s computers carries out a series of logical operations starting from one of either two states, 0 or 1, a qubit can exist in a superposition of both states. While in this delicate in-between state, a qubit should be able to simultaneously communicate with many other qubits and process multiple streams of information at a time, to quickly solve problems that would take classical computers years to process.
There are many types of qubits, some of which are engineered and others that exist naturally. Most qubits are notoriously fickle, either unable to maintain their superposition or unwilling to communicate with other qubits.
By comparison, the MIT team’s new qubit appears to be extremely robust, able to maintain a superposition between two vibrational states, even in the midst of environmental noise, for up to 10 seconds. The team believes the new vibrating qubits could be made to briefly interact, and potentially carry out tens of thousands of operations in the blink of an eye.
“We estimate it should take only a millisecond for these qubits to interact, so we can hope for 10,000 operations during that coherence time, which could be competitive with other platforms,” says Martin Zwierlein, the Thomas A. Frank Professor of Physics at MIT. “So, there is concrete hope toward making these qubits compute.”
Zwierlein is a co-author on the paper, along with lead author Thomas Hartke, Botond Oreg, and Ningyuan Jia, who are all members of MIT’s Research Laboratory of Electronics.
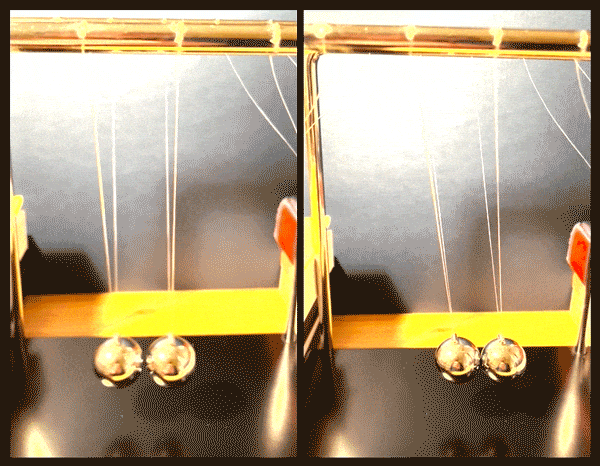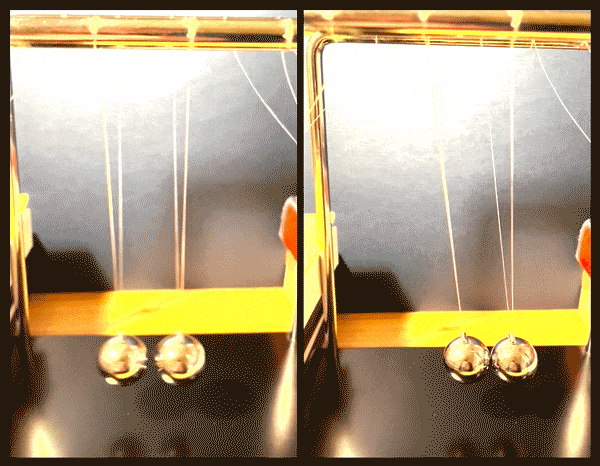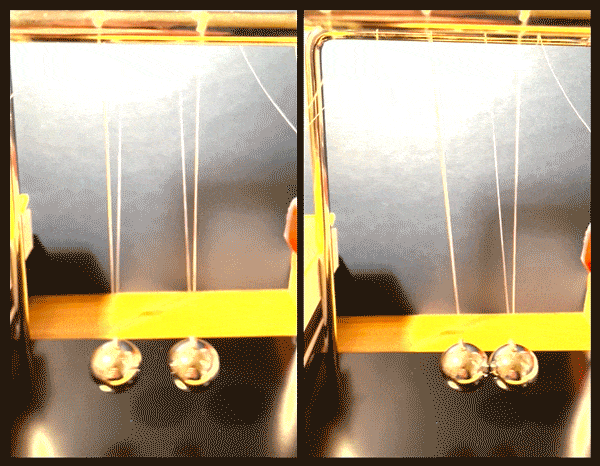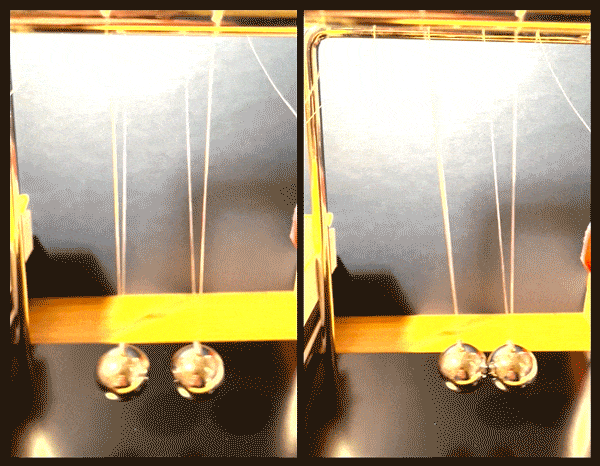
Happy accidents
The team’s discovery initially happened by chance. Zwierlein’s group studies the behavior of atoms at ultracold, super-low densities. When atoms are chilled to temperatures a millionth that of interstellar space, and isolated at densities a millionth that of air, quantum phenomena and novel states of matter can emerge.
Under these extreme conditions, Zwierlein and his colleagues were studying the behavior of fermions. A fermion is technically defined as any particle that has an odd half-integer spin, like neutrons, protons, and electrons. In practical terms, this means that fermions are prickly by nature. No two identical fermions can occupy the same quantum state — a property known as the Pauli exclusion principle. For instance, if one fermion spins up, the other must spin down.
Electrons are classic examples of fermions, and their mutual Pauli exclusion is responsible for the structure of atoms and the diversity of the periodic table of elements, along with the stability of all the matter in the universe. Fermions are also any type of atom with an odd number of elementary particles, as these atoms would also naturally repel each other.
Zwierlein’s team happened to be studying fermionic atoms of potassium-40. They cooled a cloud of fermions down to 100 nanokelvins and used a system of lasers to generate an optical lattice in which to trap the atoms. They tuned the conditions so that each well in the lattice trapped a pair of fermions. Initially, they observed that under certain conditions, each pair of fermions appeared to move in sync, like a single molecule.
To probe this vibrational state further, they gave each fermion pair a kick, then took fluorescence images of the atoms in the lattice, and saw that every so often, most squares in the lattice went dark, reflecting pairs bound in a molecule. But as they continued imaging the system, the atoms seemed to reappear, in periodic fashion, indicating that the pairs were oscillating between two quantum vibrational states.
“It’s often in experimental physics that you have some bright signal, and the next moment it goes to hell, to never come back,” Zwierlein says. “Here, it went dark, but then bright again, and repeating. That oscillation shows there is a coherent superposition evolving over time. That was a happy moment.”
“A low hum”
After further imaging and calculations, the physicists confirmed that the fermion pairs were holding a superposition of two vibrational states, simultaneously moving together, like two pendula swinging in sync, and also relative to, or against each other.
“They oscillate between these two states at about 144 hertz,” Hartke notes. “That’s a frequency you could hear, like a low hum.”
The team was able to tune this frequency, and control the vibrational states of the fermion pairs, by three orders of magnitude, by applying and varying a magnetic field, through an effect known as Feshbach resonance.
“It’s like starting with two noninteracting pendula, and by applying a magnetic field, we create a spring between them, and can vary the strength of that spring, slowly pushing the pendula apart,” Zwierlein says.
In this way, they were able to simultaneously manipulate about 400 fermion pairs. They observed that as a group, the qubits maintained a state of superposition for up to 10 seconds, before individual pairs collapsed into one or the other vibrational state.
“We show we have full control over the states of these qubits,” Zwierlein says.
To make a functional quantum computer using vibrating qubits, the team will have to find ways to also control individual fermion pairs — a problem the physicists are already close to solving. The bigger challenge will be finding a way for individual qubits to communicate with each other. For this, Zwierlein has some ideas.
“This is a system where we know we can make two qubits interact,” he says. “There are ways to lower the barrier between pairs, so that they come together, interact, then split again, for about one millisecond. So, there is a clear path toward a two-qubit gate, which is what you would need to make a quantum computer.”
Reference: “Quantum register of fermion pairs” by Thomas Hartke, Botond Oreg, Ningyuan Jia and Martin Zwierlein, 26 January 2022, Nature.
DOI: 10.1038/s41586-021-04205-8
This research was supported, in part, by the National Science Foundation, the Gordon and Betty Moore Foundation, the Vannevar Bush Faculty Fellowship, and the Alexander von Humboldt Foundation.