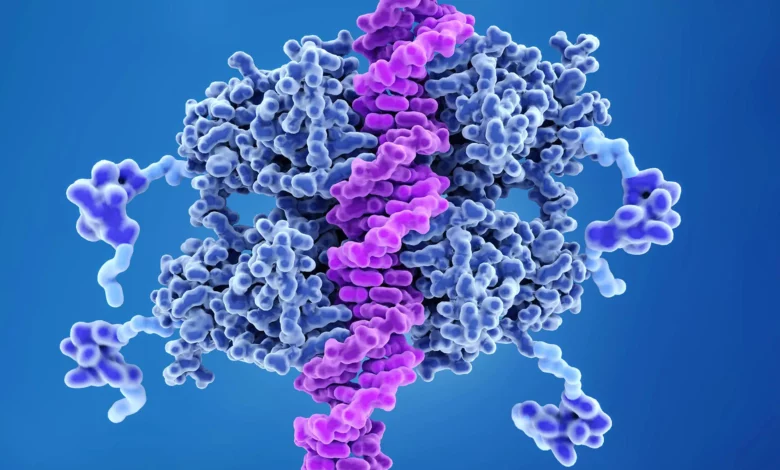
Lead Image: MIT researchers have developed a fast and precise technique using CRISPR genome-editing technology to engineer specific cancer-related mutations in mouse models, a step that could significantly advance drug development and understanding of tumor development. p53 (blue) binds to DNA (pink) to help prevent cancer formation.
With the new method, scientists can explore many cancer mutations whose roles are unknown, helping them develop new drugs that target those mutations.
MIT scientists have advanced cancer research by creating a novel method to engineer specific cancer-related mutations into mouse models using CRISPR genome-editing technology. This technique has been employed to design models of multiple mutations of the cancer-causing gene, Kras, in various organs. The fast and precise method bypasses the laborious traditional approach that took months or years to produce and analyze mice with a single cancer-linked mutation. Furthermore, the researchers hope the technique could be applied to any cancer mutation. As such, this powerful tool provides potential for identifying and testing new drugs that target these mutations and understanding their distinct effects on tumor development.
Genomic studies of cancer patients have revealed thousands of mutations linked to tumor development. However, for the vast majority of those mutations, researchers are unsure of how they contribute to cancer because there’s no easy way to study them in animal models.
In an advance that could help scientists make a dent in that long list of unexplored mutations, MIT researchers have developed a way to easily engineer specific cancer-linked mutations into mouse models.
Using this technique, which is based on CRISPR genome-editing technology, the researchers have created models of several different mutations of the cancer-causing gene Kras, in different organs. They believe this technique could also be used for nearly any other type of cancer mutation that has been identified.
Such models could help researchers identify and test new drugs that target these mutations.
“This is a remarkably powerful tool for examining the effects of essentially any mutation of interest in an intact animal, and in a fraction of the time required for earlier methods,” says Tyler Jacks, the David H. Koch Professor of Biology, a member of the Koch Institute for Integrative Cancer Research at MIT, and one of the senior authors of the new study.
Francisco Sánchez-Rivera, an assistant professor of biology at MIT and member of the Koch Institute, and David Liu, a professor in the Harvard University Department of Chemistry and Chemical Biology and a core institute member of the Broad Institute, are also senior authors of the study, which was published on May 11 in Nature Biotechnology.
Zack Ely PhD ’22, a former MIT graduate student who is now a visiting scientist at MIT, and MIT graduate student Nicolas Mathey-Andrews are the lead authors of the paper.
Faster editing
Testing cancer drugs in mouse models is an important step in determining whether they are safe and effective enough to go into human clinical trials. Over the past 20 years, researchers have used genetic engineering to create mouse models by deleting tumor suppressor genes or activating cancer-promoting genes. However, this approach is labor-intensive and requires several months or even years to produce and analyze mice with a single cancer-linked mutation.
“A graduate student can build a whole PhD around building a model for one mutation,” Ely says. “With traditional models, it would take the field decades to catch up to all of the mutations we’ve discovered with the Cancer Genome Atlas.”
In the mid-2010s, researchers began exploring the possibility of using the CRISPR genome-editing system to make cancerous mutations more easily. Some of this work occurred in Jacks’ lab, where Sánchez-Rivera (then an MIT graduate student) and his colleagues showed that they could use CRISPR to quickly and easily knock out genes that are often lost in tumors. However, while this approach makes it easy to knock out genes, it doesn’t lend itself to inserting new mutations into a gene because it relies on the cell’s DNA repair mechanisms, which tend to introduce errors.
Inspired by research from Liu’s lab at the Broad Institute, the MIT team wanted to come up with a way to perform more precise gene-editing that would allow them to make very targeted mutations to either oncogenes (genes that drive cancer) or tumor suppressors.
In 2019, Liu and colleagues reported a new version of CRISPR genome-editing called prime editing. Unlike the original version of CRISPR, which uses an enzyme called Cas9 to create double-stranded breaks in DNA, prime editing uses a modified enzyme called Cas9 nickase, which is fused to another enzyme called reverse transcriptase. This fusion enzyme cuts only one strand of the DNA helix, which avoids introducing double-stranded DNA breaks that can lead to errors when the cell repairs the DNA.
The MIT researchers designed their new mouse models by engineering the gene for the prime editor enzyme into the germline cells of the mice, which means that it will be present in every cell of the organism. The encoded prime editor enzyme allows cells to copy an RNA sequence into DNA that is incorporated into the genome. However, the prime editor gene remains silent until activated by the delivery of a specific protein called Cre recombinase.
Since the prime editing system is installed in the mouse genome, researchers can initiate tumor growth by injecting Cre recombinase into the tissue where they want a cancer mutation to be expressed, along with a guide RNA that directs Cas9 nickase to make a specific edit in the cells’ genome. The RNA guide can be designed to induce single DNA base substitutions, deletions, or additions in a specified gene, allowing the researchers to create any cancer mutation they wish.
Modeling mutations
To demonstrate the potential of this technique, the researchers engineered several different mutations into the Kras gene, which drives about 30 percent of all human cancers, including nearly all pancreatic adenocarcinomas. However, not all Kras mutations are identical. Many Kras mutations occur at a location known as G12, where the amino acid glycine is found, and depending on the mutation, this glycine can be converted into one of several different amino acids.
The researchers developed models of four different types of Kras mutations found in lung cancer: G12C, G12D, G12R, and G12A. To their surprise, they found that the tumors generated in each of these models had very different traits. For example, G12R mutations produced large, aggressive lung tumors, while G12A tumors were smaller and progressed more slowly.
Learning more about how these mutations affect tumor development differently could help researchers develop drugs that target each of the different mutations. Currently, there are only two FDA-approved drugs that target Kras mutations, and they are both specific to the G12C mutation, which accounts for about 30 percent of the Kras mutations seen in lung cancer.
The researchers also used their technique to create pancreatic organoids with several different types of mutations in the tumor suppressor gene p53, and they are now developing mouse models of these mutations. They are also working on generating models of additional Kras mutations, along with other mutations that help to confer resistance to Kras inhibitors.
“One thing that we’re excited about is looking at combinations of mutations including Kras mutations that drives tumorigenesis, along with resistance associated mutations,” Mathey-Andrews says. “We hope that will give us a handle on not just whether the mutation causes resistance, but what does a resistant tumor look like?”
The researchers have made mice with the prime editing system engineered into their genome available through a repository at the Jackson Laboratory, and they hope that other labs will begin to use this technique for their own studies of cancer mutations.
Reference: “A prime editor mouse to model a broad spectrum of somatic mutations in vivo” by Zackery A. Ely, Nicolas Mathey-Andrews, Santiago Naranjo, Samuel I. Gould, Kim L. Mercer, Gregory A. Newby, Christina M. Cabana, William M. Rideout III, Grissel Cervantes Jaramillo, Jennifer M. Khirallah, Katie Holland, Peyton B. Randolph, William A. Freed-Pastor, Jessie R. Davis, Zachary Kulstad, Peter M. K. Westcott, Lin Lin, Andrew V. Anzalone, Brendan L. Horton, Nimisha B. Pattada, Sean-Luc Shanahan, Zhongfeng Ye, Stefani Spranger, Qiaobing Xu, Francisco J. Sánchez-Rivera, David R. Liu and Tyler Jacks, 11 May 2023, Nature Biotechnology.
DOI: 10.1038/s41587-023-01783-y
The research was funded by the Ludwig Center at MIT, the National Cancer Institute, a Howard Hughes Medical Institute Hanna Grey Fellowship, the V Foundation for Cancer Research, a Koch Institute Frontier Award, the MIT Research Support Committee, a Helen Hay Whitney Postdoctoral Fellowship, the David H. Koch Graduate Fellowship Fund, the National Institutes of Health, and the Lustgarten Foundation for Pancreatic Cancer Research.
Other authors of the paper include Santiago Naranjo, Samuel Gould, Kim Mercer, Gregory Newby, Christina Cabana, William Rideout, Grissel Cervantes Jaramillo, Jennifer Khirallah, Katie Holland, Peyton Randolph, William Freed-Pastor, Jessie Davis, Zachary Kulstad, Peter Westcott, Lin Lin, Andrew Anzalone, Brendan Horton, Nimisha Pattada, Sean-Luc Shanahan, Zhongfeng Ye, Stefani Spranger, and Qiaobing Xu.